Publications
Note that these publications are ones specific to the team's
projects. Gyori's full publication list is available
on
Google Scholar.
2025
Published
Charles Tapley Hoyt, Klas Klaris, Benjamin M. Gyori
Assembly and reasoning over semantic mappings at scale for biomedical data integration
Bioinformatics, 2025
Published
Lindsey N Anderson, Charles Tapley Hoyt, Jeremy Zucker, Jeremy R. Teuton, Andrew D. McNaughton, Klas Klaris, Natasha N. Arokium-Christian, Jackson T. Warley, Zachary R. Stromberg, Benjamin M. Gyori, and Neeraj Kumar
Computational Tools and Data Integration to Accelerate Vaccine Development: Challenges, Opportunities, and Future Directions
Frontiers in Immunology, 2025
Published
Vasilevsky NA, Toro S, Matentzoglu NA, ..., Benjamin M. Gyori ... Charles Tapley Hoyt, ..., Mungall CJ, Haendel MA.
Mondo: Integrating Disease Terminology Across Communities. Genetics, 2025.
Published
Jici Jiang, Predrag Radivojac, Benjamin M. Gyori
Literature-driven extraction and computational prediction of causal statements linking genetic variants to biological processes, pathways and phenotypes
PSB, 2026
Published
Lo Surdo P, Iannuccelli M, Klas Karis, ... Benjamin M. Gyori, Livia Perfetto.
SIGNOR 4.0: the 2025 update with focus on phosphorylation data
Nucleic Acid Research, 2025
Accepted
Tong Wang, Benjamin M. Gyori, Scott T. Weiss, Giulia Menichetti, Yang-Yu Liu.
Revealing Interactions between Microbes, Metabolites, and Dietary Compounds using Genome-scale Analysis. Microbiome 2025
Preprint
Veselina Petrova, Caitlin E. Mills, ... Sepideh Fouladzadeh, ... Marian Kalocsay, Benjamin M. Gyori, Ahmet Hoke, Peter K. Sorger, Clifford J. Woolf. Multi-Kinase Inhibition of STE20 Kinases is Neuroprotective Against Chemotherapy-Induced Axon Degeneration, SSRN, 2025
Preprint
Charles Tapley Hoyt, Craig Bakker, Richard J. Callahan, Joseph Cottam, August George, Benjamin M. Gyori, Haley M. Hummel, Nathaniel Merrill, Sara Mohammad Taheri, Pruthvi Prakash Navada, Marc-Antoine Parent, Adam Rupe, Olga Vitek, Jeremy Zucker
Causal identification with Y0, arXiv, 2025
Preprint
Shawn ZK Tan, ..., Benjamin M. Gyori, Melissa A Haendel, Charles Tapley Hoyt ... David Osumi-Sutherland
The Cell Ontology in the age of single-cell omics
bioRxiv, 2025
Preprint
Vartika Tewari, Devon Kohler, Sunghyun Huh, Karen Sachs, Benjamin M. Gyori, Olga Vitek.
Simulated mass spectrometry-based protein abundances enable effective experimental design, 2025.
2024
Published
Sara Mohammad-Taheri, Pruthvi Prakash Navada, Charles Tapley Hoyt, Jeremy Zucker, Karen Sachs, Benjamin M. Gyori and Olga Vitek.
Eliater: a Python package for estimating outcomes of
perturbations in biomolecular networks.
Bioinformatics 2024.
Published
Benjamin M. Gyori and Olga Vitek
Beyond protein lists: AI-assisted interpretation of proteomic investigations in the context of evolving scientific knowledge
Nature Methods 2024.
Published
Charles Tapley Hoyt and Benjamin M. Gyori
The O3 guidelines: open data, open code, and open infrastructure for sustainable curated scientific resources
Scientific Data 2024.
Summary video: https://youtu.be/kuJsl-rRjZY
Published
Jain A, Gyori BM, Hakim S, Jain A, Sun L,
Petrova V, ..., Bunga S, ..., PK Sorger, CJ Woolf
Nociceptor-immune interactomes reveal insult-specific immune signatures of pain.
Nature Immunology, 2024
Published
Tiffany J. Callahan, Ignacio J. Tripodi, Adrianne L. Stefanski, Luca Cappelletti,
Sanya B. Taneja, Jordan M. Wyrwa, Elena Casiraghi, Nicolas A. Matentzoglu,
Justin Reese, Jonathan C. Silverstein, Charles Tapley Hoyt,
..., Lawrence E. Hunter
An open source knowledge graph ecosystem for the life sciences.
Scientific Data, 2024
2023
Published
Garano, M. A., ..., Gyori, B. M., ..., Hoyt, C. T.,
..., Robinson, P. N.
The Human Phenotype Ontology in 2024: phenotypes around the world
Nucleic Acids Research, gkad1005, 2023.
Published
Bachman JA, Gyori BM, Sorger PK
Automated assembly of molecular mechanisms at scale from text mining and curated databases
Molecular Systems Biology, e11325, 2023.
Published
Hoyt, C. T., Hoyt, A. L., and Gyori, B. M.
Prediction and Curation of Missing Biomedical Identifier Mappings with Biomappings
Bioinformatics, 39(4), btad130, 2023.
Published
Pillich RT, Chen J, Churas C, Fong D, Gyori BM, Ideker T,
Karis K, Liu SN, Ono K, Pico A, Pratt D
NDEx IQuery: a multi-method network gene set analysis leveraging the Network Data Exchange
Bioinformatics, 39(3), btad118, 2023.
Published
Lobentanzer S, Aloy P, Baumbach J, Bohar B, Danhauser K,
Doğan T, Dreo J, Dunham I, Fernandez-Torras A, Gyori BM,
Hartung M, Hoyt CT, Klein C, Korcsmaros T, Maier A,
Mann M, Ochoa D, Pareja-Lorente E, Preusse P,
Probul N, Schwikowski B, Sen B, Strauss MT, Turei D,
Ulusoy E, Wodke J, Saez-Rodriguez J
Democratising Knowledge Representation with BioCypher
Nature Biotechnology, 2023.
Published
Niarakis A, Ostaszewski M, Mazein A, ..., Gyori BM,
..., Reinhard Schneider
Drug-Target identification in COVID-19 disease mechanisms using computational systems biology approaches
Frontiers in Immunology, 2023.
2022
Published
Hoyt CT, Balk M, Callahan, TJ, Domingo-Fernandez D, Haendel MA, Hegde HB,
Himmelstein DS, Karis K, Kunze J, Lubiana T, Matentzoglu N, McMurry J, Moxon S,
Mungall CJ, Rutz A, Unni DR, Willighagen E, Winston D, & Gyori BM
Unifying the Identification of Biomedical Entities with the Bioregistry
Scientific Data, 2022.
Published
Gyori BM, Hoyt CT, Steppi A,
Gilda: biomedical entity text normalization with machine-learned disambiguation as a
service
Bioinformatics Advances, 2022.
Published
Scholten B, Guerrero Simón L, Krishnan S,
Vermeulen R, Pronk A, Gyori BM,
Bachman JA, Vlaanderen J, Stierum R.
Automated Network Assembly of Mechanistic Literature for
Informed Evidence Identification to Support Cancer Risk Assessment
Environmental Health Perspectives, 2022.
Published
Gyori BM and Hoyt CT.
PyBioPAX: biological pathway exchange in Python
Journal of Open Source Software, 2022.
Published
Balabin H, Hoyt CT, Birkenbihl C,
Gyori BM, Bachman JA,
Kodamullil AT, Ploeger PG, Hofmann-Apitius M, Domingo-Fernandez D.
STonKGs: A Sophisticated Transformer Trained on Biomedical Text and Knowledge Graphs
Bioinformatics, 2022.
Published
Balabin H, Hoyt CT,
Gyori BM, Bachman JA,
Kodamullil AT,Hofmann-Apitius M, Domingo-Fernandez D.
ProtSTonKGs: A Sophisticated Transformer Trained on Protein Sequences, Text, and Knowledge Graphs
SWAT4HCLS, 2022.
Published
N Matentzoglu, JP Balhoff, SM Bello, C Bizon, M Brush, TJ
Callahan, CG Chute, WD Duncan, CT Evelo, D Gabriel, J Graybeal, A Gray, BM Gyori, M
Haendel, H Harmse, NL Harris, I Harrow, H Hegde, AL Hoyt, CT Hoyt, ..., CJ Mungall
A Simple Standard for Sharing Ontological Mappings (SSSOM)
Database, 2022.
Published
Bonner S, Barrett IP, Ye C, Swiers R,
Engkvist O, Hoyt CT, Hamilton WL
Understanding the Performance of Knowledge Graph Embeddings in Drug Discovery
Artificial Intelligence in the Life Sciences, 2022.
Published
Doherty LM, Mills CE, Boswell SA, Liu X, Hoyt CT, Gyori
BM,
Buhrlage SJ, Sorger PK.
Integrating multi-omics data reveals function and therapeutic potential of deubiquitinating
enzymes
eLife 11:e72879, 2022.
Website: DUB portal
Published Mohammad-Taheri S,
Zucker J,
Hoyt CT,
Sachs K,
Tewari V,
Ness R,
Vitek O
Do-calculus enables causal reasoning with latent variable models
Bioinformatics, 2022.
Published
de Crécy-lagard V, de Hegedus RA, Arighi C, ..., Gyori BM, ..., Xu J
A roadmap for the functional annotation of protein families: a community perspective
Database, 2022.
Published
Rozemberczki B, Hoyt CT, Gogleva A, Grabowski P,
Karis K, Lamov A, Andriy N, Nilsson S,
Ughetto M, Wang Y, Derr T, Gyori BM.
ChemicalX: A Deep Learning Library for Drug Pair Scoring
KDD, 2022.
Published
Hungerford J, Chan YS, MacBride J,
Gyori BM, ...,
Reynolds M, Surdeanu M, Bethard S, Sharp R
Taxonomy Builder: a Data-driven and User-centric Tool for Streamlining Taxonomy Construction
NAACL HCI+NLP, 2022.
Published
Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O,
Bender A, Hoyt CT, Hamilton WL
A Review of Biomedical Datasets Relating to Drug Discovery: A Knowledge Graph Perspective
Briefings in Bioinformatics, 2022.
Published
Matentzoglu N, Goutte-Gattat D, Tan SZK, Balhoff JP, Carbon S, Caron AR,
Duncan WD, Flack JE, Haendel M, Harris NL, Hogan WR, Hoyt CT,
Jackson RC, Kim H, Kir H, Larralde M, McMurry JA, Overton JA, Peters B, ... Osumi-Sutherland D.
Ontology Development Kit: a toolkit for building, maintaining, and standardising biomedical ontologies
Database, 2022.
Preprint
Mohammad-Taheri S, Tewari V, Kapre R, Rahiminasab E, Sachs K,
Hoyt CT, Zucker J, Vitek O
Experimental design for causal query estimation in partially observed biomolecular networks
arXiv, 2022.
Preprint Bachman JA, Sorger PK, Gyori BM.
Assembling a corpus of phosphoproteomic annotations using ProtMapper to normalize site
information from databases and text mining
bioRxiv, 2022.
Preprint
Hoyt CT, Berrendorf M, Gaklin M,
Tresp V, Gyori BM.
A Unified Framework for Rank-based Evaluation Metrics for
Link Prediction in Knowledge Graphs
arXiv, 2022.
Preprint
Gaklin M, Berrendorf M, Hoyt CT
An Open Challenge for Inductive Link Prediction on Knowledge Graphs
arXiv, 2022.
2021
Published
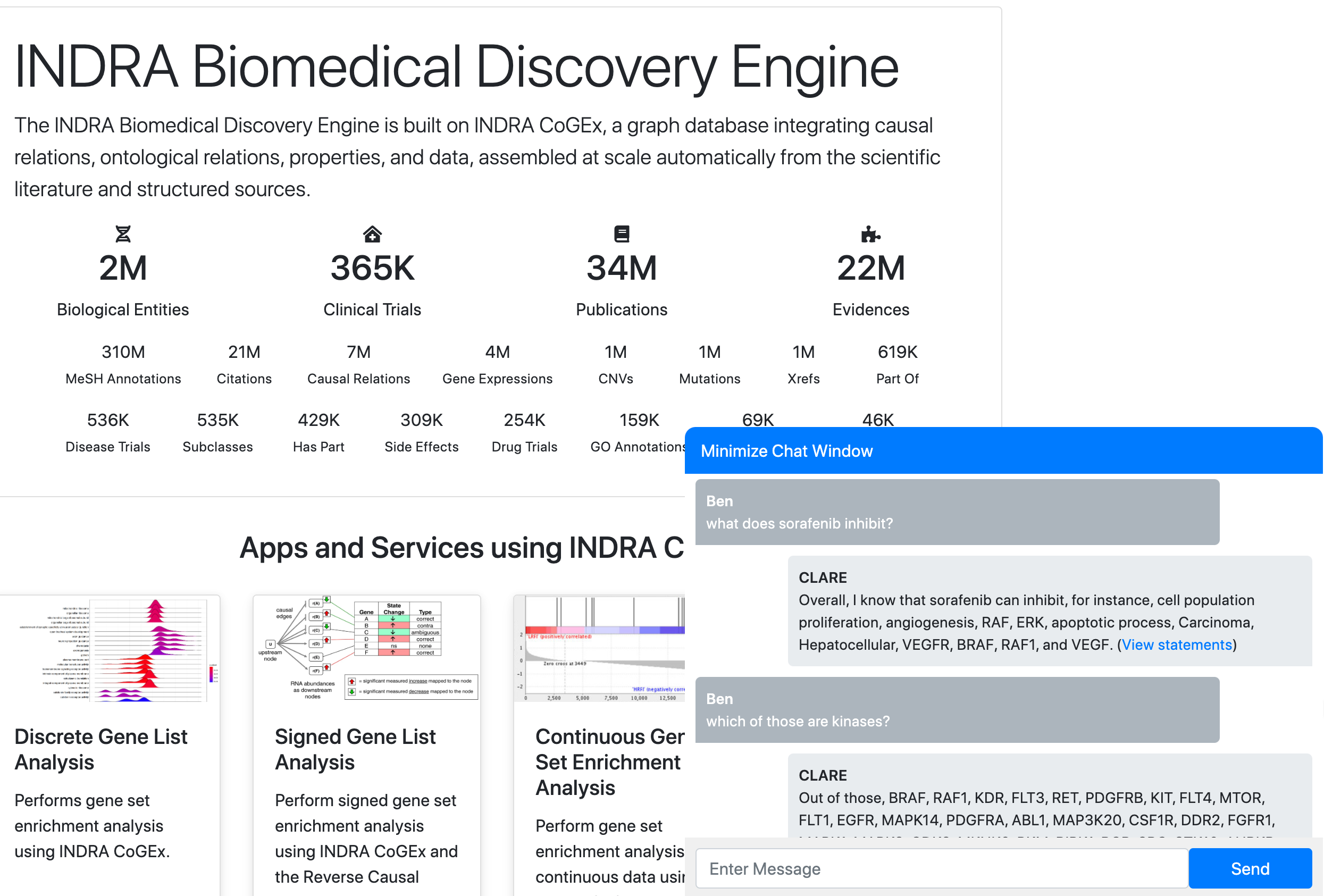
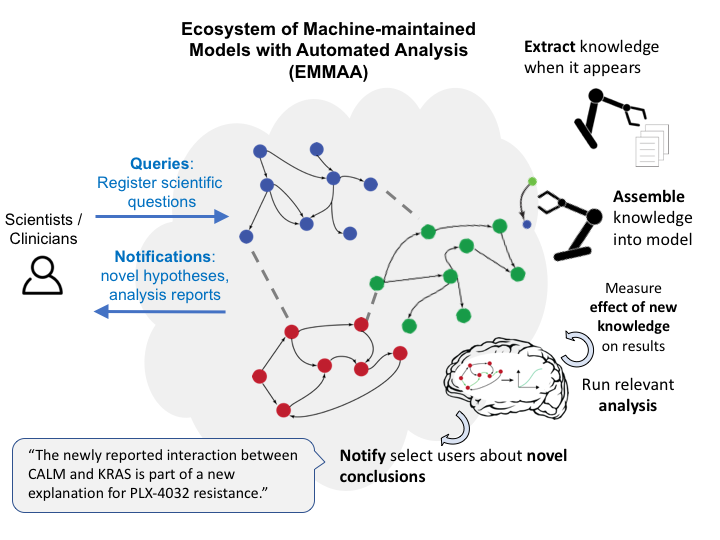
Gyori BM, Bachman JA.
From knowledge to models: automated modeling in systems and synthetic biology.
Current Opinion in Systems Biology, 2021.
Published
Ietswaart R, Gyori BM, Bachman JA, Sorger PK, Churchman S
GeneWalk identifies relevant gene functions for a biological context using network
representation learning
Genome Biology, 2021 22(51).
Published
Gyori BM, Bachman JA,
Kolusheva D.
A self-updating causal model of COVID-19 mechanisms built from the scientific literature
Proceedings of the BioCreative VII Challenge Evaluation Workshop, 2021.
Published
Wong J, Franz M, Siper MC, Fong D, Durupinar F, Dallago C, Luna A, Giorgi JM,
Rodchenkov I, Babur O, Bachman JA, Gyori BM,
Demir E, Bader G, Sander C.
Author-sourced capture of pathway knowledge in computable form using Biofactoid
eLife, 2021 10:e68292.
Published
Ostaszewski M, Niarakis A, ... Gyori BM, Bachman JA,
..., Baling R, Schneider R.
COVID-19 Disease Map, a computational knowledge repository of SARS-CoV-2 virus-host
interaction mechanisms
Molecular Systems Biology, 2021.
Preprint
Moret N, Liu C, Gyori BM, Bachman JA, Steppi A,
Taujale R, Huang LC, Hug C, Berginski M, Gomez S, Kannan N, Sorger PK.
Exploring the understudied human kinome for research and therapeutic opportunities
bioRxiv, 2021.
2020
Published
Nam KM, Gyori BM, Amethyst SV, Bates DJ, Gunawardena J
Robustness and parameter geography in post-translational modification systems
PLoS Computational Biology, 2020.
Published
Steppi A, Gyori BM, Bachman JA.
Adeft: Acromine-based Disambiguation of Entities from Text with applications to the
biomedical literature
Journal of Open Source Software, 2020.
2017-2019
Published
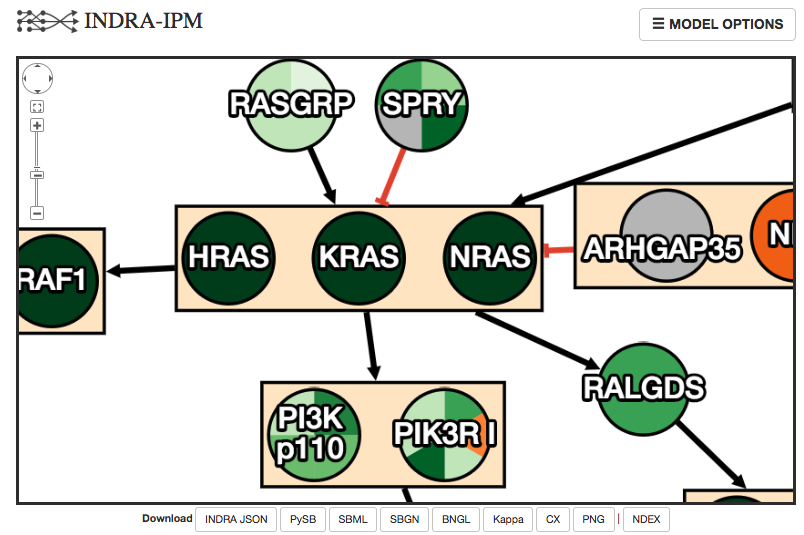
Todorov PV, Gyori BM, Bachman JA, Sorger PK.
INDRA-IPM: interactive pathway modeling
using natural language with automated assembly.
Bioinformatics, 2019.
Website: http://pathwaymap.indra.bio
Published
Sharp R, Pyarelal A, Gyori BM,
Alcock K, Laparra Egoitz,
Valenzuela-Escárcega MA, Nagesh A, Yadav V,
Bachman JA,
Tang Z, Lent H, Luo F, Paul M, Bethard S, Barnard K,
Morrison C, Surdeanu M.
Eidos, INDRA, & Delphi: From Free Text to Executable Causal Models
NAACL, 2019.
Published
Hoyt C, Domingo-Fernández D, Aldisi R, Xu L,
Kolpeja K, Spalek S, Wollert E, Bachman J, Gyori BM,
Greene P, Hofmann-Apitius M.
Re-curation and rational enrichment of knowledge graphs in
Biological Expression Language
Database, 2019.
Published
Bachman JA, Gyori BM, Sorger PK.
FamPlex: a resource for entity recognition and relationship resolution of human protein families and complexes in biomedical text mining.
BMC Bioinformatics, 2018 19(1):248.
Repository: FamPlex
Published
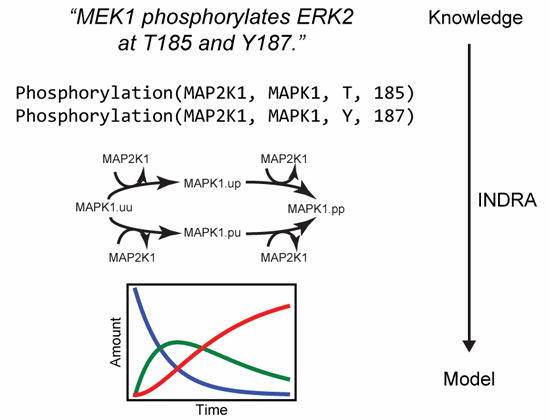
Gyori BM, Bachman JA, Subramanian K, Muhlich JL, Galescu L, Sorger PK. From word models to executable models of
signaling networks using automated assembly.
Molecular Systems Biology, 2017 13(11):954.
Summary video: https://vimeo.com/265004298